Elemental Multivariate Microbiological Analysis
EMMA: Elemental Multivariate Microbiological Analysis, which is Laser-induced breakdown spectroscopy (LIBS) used to identify bacteria, is an all-optical analytical technique that utilizes a pulsed laser and optical spectroscopy to detect and identify the elements present in a target material, including pathogenic bacteria.
It is well-known that there is an urgent and compelling need for a technology that can identify pathogens in real-time in various clinical specimens without requiring cumbersome technical preparation or genetic amplification steps. This is commonly referred to as a “rapid, point-of-care diagnostic” because the immediate diagnosis of pathogen infection allows the initiation of appropriate therapy at the point-of-care, lowering expenses and improving patient outcomes.
There is now a growing body of published work that shows the power of elemental multivariate microbiological analysis (EMMA) for rapidly identifying bacterial pathogens by measuring their unique elemental (atomic) composition (see one-page reference list at back). These new developments, coupled with recent advances in producing fieldable hardware, are strongly suggestive that a real-time EMMA-based detector technology can be developed in the near future for rapid identification of infectious disease-causing pathogens that are an ever-present threat to the public health. The speed and accuracy of this emerging technology will represent a very significant advance in medical pathogen diagnoses as well as bio-threat protection and security (important to the military, law-enforcement, and first-responder communities).
Currently, it takes approximately 2-6 hours (rapid PCR) or 24-72 hours (traditional culturing techniques) for pathogen identification technologies to obtain an accurate diagnosis. Other antibody-based identification schemes (using microchip or optical fiber technology) can only identify a single specific bacterial strain at one time and require unstable immunochemicals and other consumables. Clearly, the longer analysis times and the lack of ability to broadly identify unknown pathogens create major risks of disease spread and delay the initiation of appropriate antibiotic therapy. In our technology, pathogens are atomized by an intense laser pulse and the visible wavelength optical emission is collected and analyzed via computational multi-variate chemometric techniques.
The pathogen’s spectral fingerprint acts as a unique “bar-code” identifier which provides an immediate classification against a pre-compiled computational model of pathogens (a computerized diagnostic library). The entire process to determine whether a pathogen has been detected (yes/no) and also to identity the pathogen takes only a few seconds.
The EMMA process is presented schematically below. The technology can be made field-portable if desired, requires minimal consumables, and would be completely automated for use by medical personnel (nurses, physicians, technicians, or first responders.) Because the laser system is completely enclosed, there is no eye-safety laser risk or microbiological exposure risk to the operator. The technique requires no genetic or antigenic precursors for amplification or identification and does not require the culturing of any bacteria.
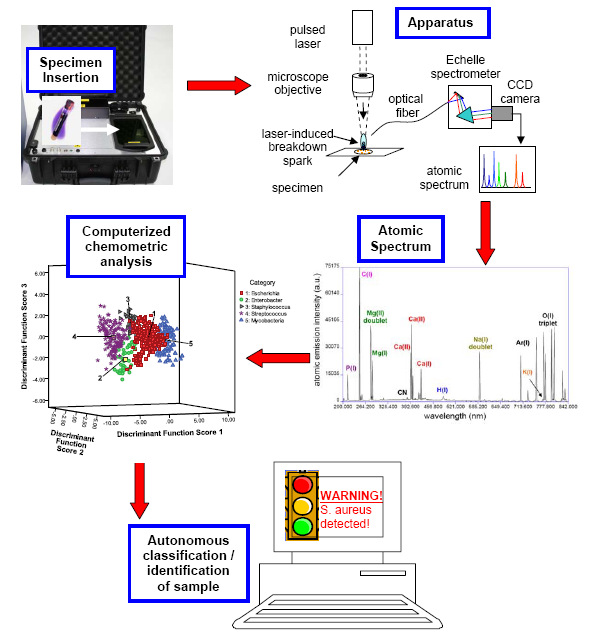
Figure 1. A schematic of how a laser-induced breakdown spectroscopy based EMMA apparatus would be used for pathogen diagnosis. (top left) A clinical specimen (which may be presented in many forms, i.e. bacterial culture, tissue biopsy, bodily fluid specimen, etc) is inserted into the portable or bench-top apparatus. Sample preparation is automated, and the laser is completely contained. (top right) The specimen is atomized by a focused (100 micron diameter) high-energy pulsed laser. Light emitted from a high-temperature spark is collected and dispersed. The atoms present in the specimen are all identified by peaks in the atomic emission spectrum (middle right), which in this case was obtained from approximately 7500 E. coli cells. The ratios of the intensities of these peaks form a “spectral fingerprint” unique to the pathogen, which can act like a bar-code to identify the pathogen. (middle left) Advanced computerized classification algorithms analyze the EMMA spectral fingerprint, and based on the specimen’s unique spectral characteristics, assign the unknown specimen to a class according to a precompiled reference library. (bottom) The results of the diagnostic analysis are conveyed in real-time to the operator (physician, technician, etc.). Because the fingerprint is obtained easily and quickly, in under one second, it can also be used to quantify changes in the specimen with time.
Things that make a LIBS-based technology unique
- lack of complicated sample preparation
- no expertise required
- no genetic or antigenic precursors (consumables) necessary
- speed / portability / durability (ruggedness)
- “rapid point-of-care diagnostic…”
- same technology / hardware useful for explosives, chemical, other threats (CBRNE capable)
- capability of sensor fusion
We have published multiple papers detailing the experiments that have demonstrated the viability – including sufficient sensitivity and specificity and a realistically low limit of detection – of the EMMA technique. The papers are all provided in our "Papers" section and the important conclusions are presented here.
- A rapid discrimination of bacterial LIBS signatures from other biotypes, including common contaminants such as yeasts or molds is possible.
- Discrimination of E. coli strains, particularly the ability to discriminate the pathogenic enterohemmorhagic E. coli O157:H7 strain from other non-pathogenic strains is possible. EMMA strain differentiation was also demonstrated by Multari et al. in Staphylococcus aureus, with several MRSA strains being differentiated on the basis of their is discussed in detail later in this Review.
- Our studies to date show that bacterial identification appears to be independent of the nutrient medium upon which the bacteria were grown (i.e. nutrient rich tryptic soy agar, or broth, blood agar medium, McConkey agar, chocolate agar, etc). This result has been confirmed by Marcos-Martinez et al. on three similar growth media.
- Bacterial LIBS spectra do not change significantly with time as the bacterial reproduction progresses from lag phase, to log phase to the stationary phase or they lie dormant (without multiplying) on a nutrient-free abiotic surface in the stationary or death phase. This is necessary for accurate identification and detection of surface contamination.
- Bacterial LIBS spectra can be easily obtained from killed (via autoclaving) or inactivated (via bactericidal UV light) specimens, and such treatment (which renders the specimen completely safe for handling) does not decrease identification specificity and does not decrease LIBS spectral intensity.
- Intensity of the LIBS spectrum is linearly dependent on cell number (bacterial titer), but the specificity is not dependent on cell number.
- Accurate EMMA identification can be made from as few as 1500 cells, although 7500 cells per “spectral fingerprint” were used to provide publication high signal to noise results in our publications. LIBS spectra have been obtained from single bacterial spores by other groups in the past.
- All species of bacteria tested to date have possessed unique atomic compositions allowing a LIBS-based identification of unknown bacterial specimens. In a blind external validation test of a five genus model composed of two Staphylococcus species (aureus and saprophyticus), two Streptococcus species (viridans and mutans), three strains of Mycobacterium smegmatis, a single strain of Enterobacter cloacae, and five strains of Escherichia coli EMMA sensitivities with our current apparatus were in the 85% range and specificities were upwards of 95%.
- Bacteria in mixed samples are identifiable. The dominant or majority bacterial component of a two-component bacterial mixture is reliably identified provided it comprises 70% of the mixture or more. Mixtures caused by specimen contamination in clinical settings will not affect EMMA sensitivity.
- Bacteria can be identified when specimens are obtained from clinical samples (e.g. sterile urine containing organic and inorganic solutes) without washing.
- Bacteria can be discriminated in air, argon, or helium environments, although argon and helium offer distinct advantages.
- Bacterial LIBS signatures appear to be correlated with bacterial membrane biochemistry (for Gram-negative bacteria) and may be related to serotype identification.
- The bacterial LIBS spectrum for a given species is stable and does not change with time (experiments conducted on the same E. coli strain over the course of multiple years).
EMMA is already a demonstrated technology for the identification of pathogens and a proven technology for the detection and identification of a variety of chemical, biological, and explosive threats due considerably in part to investments of the US Army, Defense Threat Reduction Agency (DTRA), and other US Department of Defense agencies. Leveraging the expertise that already exists in the development of field-deployable hardware and software, we now aim to develop a field-deployable point-of-care diagnostic EMMA instrument. This will require the accomplishment and integration of four important goals:
- Construction of a library of LIBS fingerprints from known pathogens. Work will begin with multiple strains of medically-important and relevant organisms. The availability of a large number of pathogen samples at the WSU medical complex will greatly speed and assist this goal.
- Increasing the EMMA limits of detection to the level of clinical specimens (titers of 10’s to 100’s of cells), while improving sensitivity and specificity above 95%.
- Development of sample preparation protocols for human specimens of blood, urine, and sputum – the front end of the EMMA package.
- Integrating the new front end into existing technology and hardware to yield a commercial product that will be available for use in clinical settings.
During LIBS, a short pulse of laser light is focused to a small spot on a bacteria-containing target (blood, sputum, etc.) which creates a high-temperature (10,000–20,000 K) micro-plasma within the focal region of the laser. During this process, the sample illuminated by the laser is completely vaporized (“ablated”). The sample is reduced to its constituent atomic components, which are entrained in the micro-plasma plume. A careful spectroscopic analysis of the light emitted from this plasma plume yields identifiable emission lines only from those elements that were present in the target (Cremers and Radziemski, 2006; Miziolek et al. 2006). The positive identification of many elemental lines within the emission spectrum then provides an immediate and unique spectral “fingerprint” which positively identifies the bacteria in the sample. This sample could be a bacterial colony, a solid surface, a vial of bacteria-containing water, or any clinical sample (e.g. sputum, blood, etc.). Any target material which can be illuminated by the laser can theoretically be tested to reveal the presence and identity of a pathogen. Beginning in 2003, studies began to show that the ability of LIBS to rapidly (within seconds or minutes) detect harmful pathogens including those which cannot be cultured within a reasonable amount of time or cannot be cultured at all could offer a radically new paradigm to the health sciences for the detection, identification, and control of infectious diseases.
A MICROBIOLOGICAL LIBS BIBLIOGRAPHY
Provided here is a list of recent papers (with the host institutions listed) that describe the use of LIBS to identify microbiological specimens. LIBS bibliography
